Synthetic Pumping Test - 2 aquifers#
import matplotlib.pyplot as plt
import numpy as np
from timflow import transient as tft
plt.rcParams["figure.figsize"] = (6, 4)
Head data is generated for a pumping test in a two-aquifer model. The well starts pumping at time \(t=0\) with a discharge \(Q=800\) m\(^3\)/d. The head is measured in an observation well 10 m from the pumping well. The thickness of the aquifer is 20 m. Questions:
Determine the optimal values of the hydraulic conductivity and specific storage coefficient of the aquifer when the aquifer is approximated as confined. Use a least squares approach and make use of the
fminfunction ofscipy.optimizeto find the optimal values. Plot the data with dots and the best-fit model in one graph. Print the optimal values of \(k\) and \(S_s\) to the screen as well as the root mean squared error of the residuals.Repeat Question 1 but now approximate the aquifer as semi-confined. Plot the data with dots and the best-fit model in one graph. Print to the screen the optimal values of \(k\), \(S_s\) and \(c\) to the screen as well as the root mean squared error of the residuals. Is the semi-cofined model a better fit than the confined model?
def generate_data():
# 2 layer model with some random error
ml = tft.ModelMaq(
kaq=[10, 20],
z=[0, -20, -22, -42],
c=[1000],
Saq=[0.0002, 0.0001],
tmin=0.001,
tmax=100,
)
tft.Well(ml, 0, 0, rw=0.3, tsandQ=[(0, 800)])
ml.solve()
t = np.logspace(-2, 1, 100)
h = ml.head(10, 0, t)
plt.figure()
r = 0.01 * rnd.random(100)
n = np.zeros_like(r)
# alpha = 0.8
for i in range(1, len(n)):
n[i] = 0.8 * n[i - 1] + r[i]
ho = h[0] + n
plt.plot(t, ho, ".")
data = np.zeros((len(ho), 2))
data[:, 0] = t
data[:, 1] = ho
# np.savetxt('pumpingtestdata.txt', data, fmt='%2.3f', header='time (d), head (m)')
return data
rnd = np.random.default_rng(11)
data = generate_data()
to = data[:, 0]
ho = data[:, 1]
self.neq 1
solution complete
def func(p, to=to, ho=ho, returnmodel=False):
k = p[0]
S = p[1]
ml = tft.ModelMaq(kaq=k, z=[0, -20], Saq=S, tmin=0.001, tmax=100)
tft.Well(ml, 0, 0, rw=0.3, tsandQ=[(0, 800)])
ml.solve(silent=True)
if returnmodel:
return ml
h = ml.head(10, 0, to)
return np.sum((h[0] - ho) ** 2)
from scipy.optimize import fmin
lsopt = fmin(func, [10, 1e-4])
print("optimal parameters:", lsopt)
print("rmse:", np.sqrt(func(lsopt) / len(ho)))
Optimization terminated successfully.
Current function value: 0.251681
Iterations: 41
Function evaluations: 85
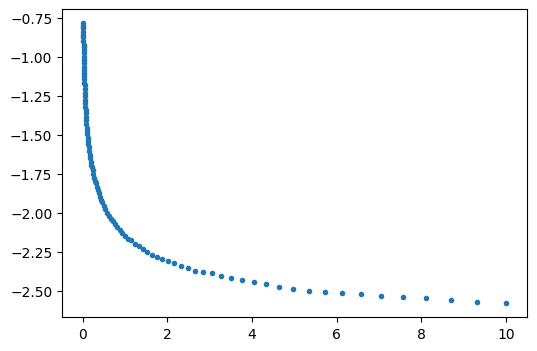
optimal parameters: [1.16063293e+01 1.27878469e-04]
rmse: 0.05016784528826497
ml = func(lsopt, returnmodel=True)
plt.figure()
plt.plot(data[:, 0], data[:, 1], ".", label="observed")
hm = ml.head(10, 0, to)
plt.plot(to, hm[0], "r", label="modeled")
plt.legend()
plt.xlabel("time (d)")
plt.ylabel("head (m)")
Text(0, 0.5, 'head (m)')
cal = tft.Calibrate(ml)
cal.set_parameter(name="kaq", layers=0, initial=10, pmin=0.1, pmax=1000)
cal.set_parameter(name="Saq", layers=0, initial=1e-4, pmin=1e-5, pmax=1e-3)
cal.series(name="obs1", x=10, y=0, layer=0, t=to, h=ho)
cal.fit(report=False)
print("rmse:", cal.rmse())
.................
...
Fit succeeded.
rmse: 0.050167851002538885
cal.parameters
| layers | optimal | std | perc_std | pmin | pmax | initial | inhoms | parray | |
|---|---|---|---|---|---|---|---|---|---|
| kaq_0_0 | 0 | 11.606190 | 0.118773 | 1.023363 | 0.10000 | 1000.000 | 10.0000 | None | [[11.606189706450678]] |
| Saq_0_0 | 0 | 0.000128 | 0.000007 | 5.559295 | 0.00001 | 0.001 | 0.0001 | None | [[0.00012788749251955707]] |
Model as semi-confined#
def func2(p, to=to, ho=ho, returnmodel=False):
k = p[0]
S = p[1]
c = p[2]
ml = tft.ModelMaq(
kaq=k, z=[2, 0, -20], Saq=S, c=c, topboundary="semi", tmin=0.001, tmax=100
)
tft.Well(ml, 0, 0, rw=0.3, tsandQ=[(0, 800)])
ml.solve(silent=True)
if returnmodel:
return ml
h = ml.head(10, 0, to)
return np.sum((h[0] - ho) ** 2)
lsopt2 = fmin(func2, [10, 1e-4, 1000])
print("optimal parameters:", lsopt2)
print("rmse:", np.sqrt(func2(lsopt2) / len(ho)))
Optimization terminated successfully.
Current function value: 0.004826
Iterations: 122
Function evaluations: 239
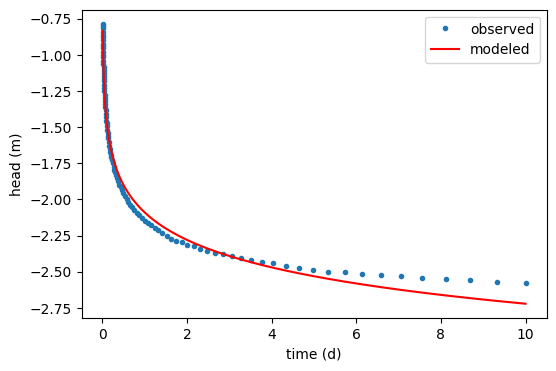
optimal parameters: [1.02193607e+01 2.01296811e-04 1.56256292e+03]
rmse: 0.006947175894895784
ml = func2(lsopt2, returnmodel=True)
plt.figure()
plt.plot(data[:, 0], data[:, 1], ".", label="observed")
hm = ml.head(10, 0, to)
plt.plot(to, hm[0], "r", label="modeled")
plt.legend()
plt.xlabel("time (d)")
plt.ylabel("head (m)")
Text(0, 0.5, 'head (m)')
ml = tft.ModelMaq(
kaq=10, z=[2, 0, -20], Saq=1e-4, c=1000, topboundary="semi", tmin=0.001, tmax=100
)
w = tft.Well(ml, 0, 0, rw=0.3, tsandQ=[(0, 800)])
ml.solve(silent=True)
cal = tft.Calibrate(ml)
cal.set_parameter(name="kaq", layers=0, initial=10)
cal.set_parameter(name="Saq", layers=0, initial=1e-4)
cal.set_parameter(name="c", layers=0, initial=1000)
cal.series(name="obs1", x=10, y=0, layer=0, t=to, h=ho)
cal.fit(report=False)
cal.parameters
..
...............
....
...............
..
Fit succeeded.
| layers | optimal | std | perc_std | pmin | pmax | initial | inhoms | parray | |
|---|---|---|---|---|---|---|---|---|---|
| kaq_0_0 | 0 | 10.219389 | 0.023068 | 0.225723 | -inf | inf | 10.0000 | None | [[10.219389301148812]] |
| Saq_0_0 | 0 | 0.000201 | 0.000002 | 0.880853 | -inf | inf | 0.0001 | None | [[0.00020129637392494947]] |
| c_0_0 | 0 | 1562.605654 | 37.890827 | 2.424849 | -inf | inf | 1000.0000 | None | [[1562.6056541553019]] |
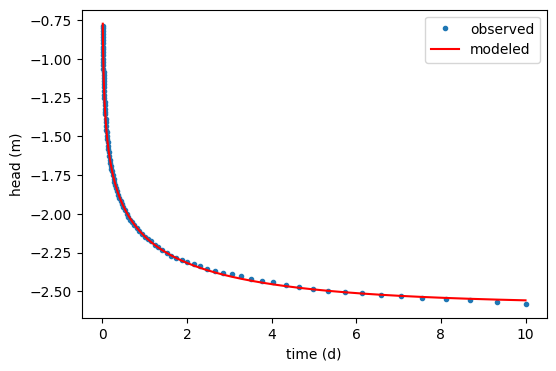
cal.rmse(), ml.aq.kaq
(np.float64(0.00694718207260787), array([10.2193893]))
plt.figure()
plt.plot(data[:, 0], data[:, 1], ".", label="observed")
hm = ml.head(10, 0, to)
plt.plot(to, hm[0], "r", label="modeled")
plt.legend()
plt.xlabel("time (d)")
plt.ylabel("head (m)")
Text(0, 0.5, 'head (m)')